Coming Soon
Includes all controls.
Cell-free Mutation Detection
The KRAS and NRAS genes encode small GTPases that mediate growth factor receptor signaling to downstream effectors that play a role in cell proliferation, survival, and differentiation. KRAS mutations have been commonly found in several types of human malignancies including metastatic colorectal cancer (CRC), lung adenocarcinoma, and thyroid carcinoma. NRAS mutations are found in ~1-6% of CRCs and occur in melanoma and thyroid carcinoma as well.
Several studies have demonstrated that tumors carrying any of these mutant forms of the KRAS gene are less likely to respond to anti-EGFR antibody therapy. [1,2] The American Society of Clinical Oncology (ASCO) released its first Provisional Clinical Opinion (PCO) suggesting that all patients to be administered anti-EGFR monoclonal antibody therapy should be screened for KRAS mutations. [3] Additionally, studies have shown that metastatic colorectal tumors carrying mutations in exons 2, 3 and 4 of KRAS and NRAS are less responsive to EGFR inhibitors. [4] These findings strongly suggest that screening for both KRAS and NRAS mutations is necessary to more accurately identify patients who will not respond to anti-EGFR therapy.
A number of recent studies focus on the significance of KRAS mutations detected in cell-free DNA. Although liquid biopsy for RAS mutations is still investigational, a meta-analysis shows that KRAS mutations in cell-free DNA of cancer patient seem to act as a survival prognostic biomarker. [5]
The ctRAS Mutation Detection kit is a non-invasive, ultra-sensitive test that selectively amplifies and detects mutations in exons 2, 3, and 4 in KRAS and NRAS from cell-free DNA derived from human plasma.
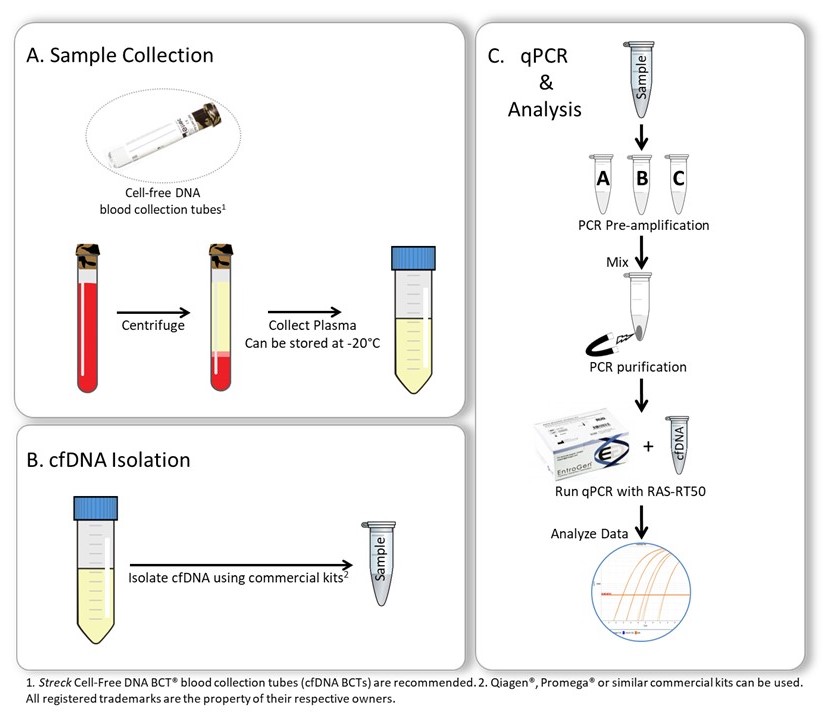
Testing Procedure
The ctRAS Mutation Detection Kit is a real-time polymerase chain reaction (PCR)-based assay that uses wildtype blockers to selectively amplify mutant circulating tumor DNA (ctDNA) in 3 separate reactions. The 3 reactions are subsequently mixed and cleaned up using provided PCR clean-up beads. Following elution, the sample is analyzed by EntroGen’s RAS Mutation Screening Panel (RAS-RT50), an allele-specific primer based real-time PCR assay.
The testing procedure involves 5 simple steps:
- Isolation of DNA from plasma
- Selective amplification of mutant ctDNA and bead-based clean up
- PCR setup with the RAS Mutation Screening Panel reagents
- Amplification and detection using a real-time PCR instrument
- Data analysis
Equipment and Materials
The ctRAS Mutation Detection kit requires a real-time PCR instrument capable of detecting FAM and VIC fluorescent probes simultaneously.
The kit includes reagents required for the mutant ctDNA selective amplification and subsequent PCR amplification/detection, as well as validated reaction controls. One kit provides reagents for up to 50 tests including all samples and controls. Columns and reagents for cell-free DNA isolation are not included.
Intended Use:
USA: Available for research use only (RUO). Not for use in diagnostic procedures.
Europe: Available for research use only (RUO). Not for use in diagnostic procedures (CE-IVD pending).
References:
- Andreyev, H.J., et al. (2001) Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 85, 692-696
- Esteller, M., et al. (2001) K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol 19, 299-304
- Allegra, C.J., et al. (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27, 2091-2096
- Douillard, JY. et al. (2013) Panitumumab-FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. N Engl J Med 369, 1023-1034
- Zhuang, R, Li S, Li Q, Guo X, Shen F, Sun H, et al. (2017) The prognostic value of KRAS mutation by cell-free DNA in cancer patients: A systematic review and meta-analysis. PLoS One 12(8): e0182562.